Dostupne verzije
| Verzija | Modul | Paralelna okolina |
|---|
| 22.04 | rapids/22.04 | gpusingle |
Korištenje
Dva su načina korištenja dostupna:
- jedan GPU
- više GPU-ova na jednom čvoru
U prvom slučaju nisu potrebne dodatne postavke, već se rapids knjižnice zovu direktno.
U drugom slučaju je potrebno instancirati lokalni klaster putem LocalCUDACluster naredbe iz knjižnice dask-cuda (primjer se nalazi ispod).
from dask.distributed import Client
from dask_cuda import LocalCUDACluster
# pokreni klaster i spoji se klijentom
cluster = LocalCUDACluster(
interface="ib0",
protocol="ucx",
)
client = Client(cluster)
# izradi proračun
...
# ugasi workere i zatvori klijenta
workers = client.get_scheduler_info()['workers']
client.retire_workers(workers=workers)
client.close()
bla
Postavke
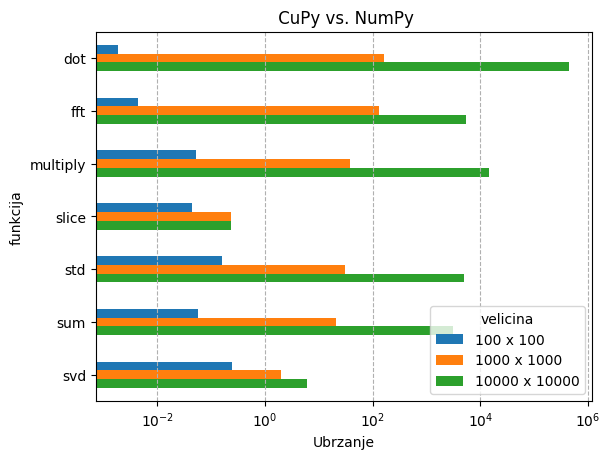
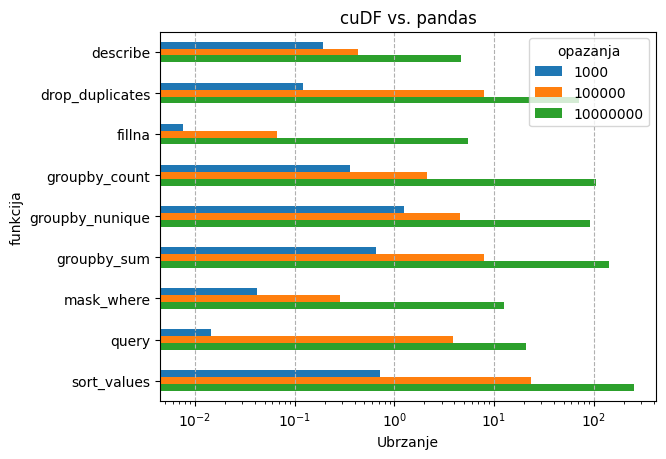
Ispod se nalazi rezultati nekih tipičnih aplikacija na jednoj GPU jezgri:
- Matrične operacije - cuPy vs. NumPy
- DataFrame operacije - cuDF vs. pandas
- K-means clustering - cuML vs. sklearn
U slučaju cuPy-a, matrične operacije su se testirale na matricama veličina:
- 100 x 100 - 80 Kb
- 1000 x 1000 - 8 Mb
- 10000 x 10000 - 0.8 Gb
U slučaju cuDF-a, dataframe operacije su se testirale na podacima strukture:
- 10 kategorija
- 10 varijabli
- 10**3, 10**5 i 10**7 opažanja
U slučaju cuML-a, klasteriranje se testiralo na podacima strukture:
- 2 varijable
- 5 klastera
- 10**8 opažanja
Kod
cuPy
#$ -cwd
#$ -o output/
#$ -e output/
#$ -pe gpu 1
# module
module load rapids/22.04
# python
cuda-wrapper.sh python cupy_test.py
import timeit
import cupy as cp
import numpy as np
def print_time(function, message):
n = 10
t = timeit.timeit( lambda: function(), number=n )
print( '%s %0.10f' % ( message, t/n ) )
if __name__ == '__main__':
# define matrix sizes
sizes = [ 100, 1000, 10000 ]
# create matrices
np_matrices = [ np.random.random((size, size)) for size in sizes ]
cp_matrices = [ cp.random.random((size, size)) for size in sizes ]
for i, (np_matrix, cp_matrix) in enumerate(zip(np_matrices, cp_matrices)):
# print nbytes
print( 'np %i nbytes %s' % ( np_matrix.shape[0], np_matrix.nbytes ) )
print( 'cp %i nbytes %s' % ( cp_matrix.shape[0], cp_matrix.nbytes ) )
# fft
print_time( lambda: np.fft.fft2(np_matrix), 'np %s fft' % np_matrix.shape[0] )
print_time( lambda: cp.fft.fft2(cp_matrix), 'cp %s fft' % cp_matrix.shape[0] )
# sum
print_time( lambda: np_matrix.sum(), 'np %s sum' % np_matrix.shape[0] )
print_time( lambda: cp_matrix.sum(), 'cp %s sum' % cp_matrix.shape[0] )
# std
print_time( lambda: np_matrix.std(), 'np %s std' % np_matrix.shape[0] )
print_time( lambda: cp_matrix.std(), 'cp %s std' % cp_matrix.shape[0] )
# elementwise
print_time( lambda: np.multiply(np_matrix, np_matrix), 'np %s multiply' % np_matrix.shape[0] )
print_time( lambda: cp.multiply(cp_matrix, cp_matrix), 'cp %s multiply' % cp_matrix.shape[0] )
# dot
print_time( lambda: np_matrix.dot(np_matrix), 'np %s dot' % np_matrix.shape[0] )
print_time( lambda: cp_matrix.dot(cp_matrix), 'cp %s dot' % cp_matrix.shape[0] )
# slice
print_time( lambda: np_matrix[::10], 'np %s slice' % np_matrix.shape[0] )
print_time( lambda: cp_matrix[::10], 'cp %s slice' % cp_matrix.shape[0] )
# svd
print_time( lambda: np.linalg.svd(np_matrix), 'np %s svd' % np_matrix.shape[0] )
print_time( lambda: np.linalg.svd(cp_matrix), 'cp %s svd' % cp_matrix.shape[0] )
cuDF
#$ -cwd
#$ -o output/
#$ -e output/
#$ -pe gpu 1
# module
module load rapids/22.04
# run python
cuda-wrapper.sh python cudf_test.py
import time
import cudf
import cupy as cp
import numpy as np
import pandas as pd
import random
def print_time(function, message):
start = time.time()
out = function()
end = time.time()
print( '%s %0.4e' % ( message, end-start ) )
return out
if __name__ == '__main__':
# change number of rows
ncat = 10
nobs = 10
rows = [ 10**n for n in range(3, 8, 2) ]
for nrows in rows:
# fake dataset
cdf = cudf.DataFrame()
cats = cudf.DataFrame(
data = cp.random.randint(0, ncat, (nrows, ncat)),
columns = [ 'c%i' % i for i in range(ncat) ],
)
cdf = cudf.concat([ cdf, cats ], axis=1)
del cats
obs = cudf.DataFrame(
data = cp.random.random(size=(nrows, nobs)),
columns = [ 'o%i' % i for i in range(nobs) ],
)
cdf = cudf.concat([ cdf, obs ], axis=1)
del obs
pdf = cdf.to_pandas()
print('cdf %i nbytes %i' % (nrows, cdf.memory_usage().sum()))
print('pdf %i nbytes %i' % (nrows, pdf.memory_usage().sum()))
# groupby
cats = [ column for column in cdf.columns if 'c' in column ]
print_time( lambda: cdf.groupby(cats).sum(), 'cdf %i groupby_sum' % nrows )
print_time( lambda: pdf.groupby(cats).sum(), 'pdf %i groupby_sum' % nrows )
print_time( lambda: cdf.groupby(cats).count(), 'cdf %i groupby_count' % nrows )
print_time( lambda: pdf.groupby(cats).count(), 'pdf %i groupby_count' % nrows )
print_time( lambda: cdf.groupby(cats).nunique(), 'cdf %i groupby_nunique' % nrows )
print_time( lambda: pdf.groupby(cats).nunique(), 'pdf %i groupby_nunique' % nrows )
# describe
print_time( lambda: cdf.describe(), 'cdf %i describe' % nrows )
print_time( lambda: pdf.describe(), 'pdf %i describe' % nrows )
# sort
print_time( lambda: cdf.sort_values(by=list(cdf.columns), axis=0), 'cdf %i sort_values' % nrows )
print_time( lambda: pdf.sort_values(by=list(pdf.columns), axis=0), 'pdf %i sort_values' % nrows )
# drop_duplicates
print_time( lambda: cdf.drop_duplicates(), 'cdf %i drop_duplicates' % nrows )
print_time( lambda: pdf.drop_duplicates(), 'pdf %i drop_duplicates' % nrows )
# mask_where
ind = [ 'o' in col for col in cdf.columns ]
cdf = print_time( lambda: cdf.mask( (cdf > 0.05) & ind, np.nan ), 'cdf %i mask_where' % nrows )
pdf = print_time( lambda: pdf.mask( (pdf > 0.05) & ind, np.nan ), 'pdf %i mask_where' % nrows )
# query
query = ' | '.join([ '( o%i > 0 )' % i for i in range(10) ])
cdf = print_time( lambda: cdf.query( query ), 'cdf %i query' % nrows )
pdf = print_time( lambda: pdf.query( query ), 'pdf %i query' % nrows )
# fillna
cdf = print_time( lambda: cdf.fillna(-1), 'cdf %i fillna' % nrows )
pdf = print_time( lambda: pdf.fillna(-1), 'pdf %i fillna' % nrows )
K-means
#$ -cwd
#$ -o output/
#$ -e output/
#$ -pe gpu 1
# module
module load rapids/22.04
# run python
cuda-wrapper.sh python kmeans_demo.py
#
# https://github.com/rapidsai/cuml/blob/branch-21.12/notebooks/kmeans_demo.ipynb
#
import sys
import cudf
import cupy
import time
import matplotlib.pyplot as plt
from cuml.cluster import KMeans as cuKMeans
from cuml.datasets import make_blobs
from sklearn.cluster import KMeans as skKMeans
from sklearn.metrics import adjusted_rand_score
# time_it
def time_it(command, message):
start = time.time()
out = command()
end = time.time()
print('%-20s %s' % ( message, end-start ))
return out
if __name__ == '__main__':
# parameters
n_samples = 10**8
n_features = 2
n_clusters = 5
random_state = 0
# generate data
device_data, device_labels = make_blobs(
n_samples=n_samples,
n_features=n_features,
centers=n_clusters,
cluster_std=0.1,
)
device_data = cudf.DataFrame(device_data)
device_labels = cudf.Series(device_labels)
host_data = device_data.to_pandas()
host_labels = device_labels.to_pandas()
# scikit
kmeans_sk = skKMeans(
init="k-means++",
n_clusters=n_clusters,
random_state=random_state,
)
time_it( lambda: kmeans_sk.fit(host_data), 'sklearn fit' )
# cuml
kmeans_cuml = cuKMeans(
init="k-means||",
n_clusters=n_clusters,
oversampling_factor=40,
random_state=random_state,
)
time_it( lambda: kmeans_cuml.fit(device_data), 'cuml fit' )
# compare
cuml_score = adjusted_rand_score(host_labels, kmeans_cuml.labels_.to_numpy())
sk_score = adjusted_rand_score(host_labels, kmeans_sk.labels_)
threshold = 1e-4
passed = ( cuml_score - sk_score ) < threshold
print('compare kmeans: cuml vs sklearn labels_ are ' + ('equal' if passed else 'not equal'))
Rezultati
cuPy
cuDF
K-means
| knjižnica | funkcija | Vrijeme [s] |
|---|
| sklearn | sklearn.cluster.KMeans.fit | 1702.17 |
| sklearn.cluster.KMeans.predict | 55.73 |
| cuML | cuML.cluster.KMeans.fit | 7.43 |
| cuML.cluster.KMeans.predict | 1.58 |

